All News
Patro Awarded $400K to Enhance Open-Source Gene Expression Tools
The concept of gene expression analysis might compare to an orchestra’s conductor—a guiding force for researchers intent on deciphering the genomic melodies within our cells. From unraveling the mysteries of diseases to better understanding human development, this scientific method significantly advances biomedical research by shedding light on life’s inner workings.
To further research and scholarship that is focused on gene expression analysis, the Chan Zuckerberg Initiative (CZI) has awarded a $400,000 grant to a team of University of Maryland researchers. CZI is a philanthropic effort launched in 2015 by Meta founder Mark Zuckerberg and his wife, Priscilla Chan.
The award supports efforts by Rob Patro, an associate professor of computer science, in unifying and enhancing open-source transcriptomics tools—freely available software programs designed to analyze RNA transcripts, enabling researchers to efficiently study gene expression and regulation.
Patro, who has an appointment in the University of Maryland Institute for Advanced Computer Studies (UMIACS), says he is honored to receive this grant, saying it highlights the value of his lab’s commitment to furthering open science methodology and expanding access to open-source, practical and robust scientific software.
“This award will improve upon the core algorithms and data structures of software tools that have been used in thousands of gene expression analysis studies to date, increasing performance and reducing runtime and resource requirements and hence, costs,” says Patro, who is a core member of the Center for Bioinformatics and Computational Biology. “At the same time, it will build new bridges of interoperability between popular tools, accelerating existing pipelines and providing scientists with more choices in how best to analyze their data.”
One primary goal, Patro says, is to enhance the use and impact of a popular open-source tool known as the STAR aligner, which uses a suffix array—a data structure for efficiently handling large sequences of DNA, RNA or protein data—to align genomic sequencing data.
Despite the suffix array’s long history and optimization for efficient construction, it hasn’t fully utilized modern hardware capabilities, Patro says.
Working closely with Jamshed Khan, who just completed his fifth-year as a computer science doctoral student—and in collaboration with UMIACS colleagues Laxman Dhulipala and Erin Molloy as well as graduate student Tobias Rubel—the UMD team has developed a new parallel algorithm for suffix array construction that outperforms existing methods, building the index faster while simultaneously reducing memory usage. By bringing this novel algorithm and other enhancements to the STAR aligner, the team hopes to improve gene analysis tool’s performance.
Another focus is to integrate various transcriptome analysis tools. The UMD researchers will create a high-bandwidth connection between STAR and their own Salmon tool for transcript quantification from bulk RNA sequencing data, reducing runtime and resource usage. A similar integration will be built between STAR and their alevin-fry tool for single-cell gene expression analysis.
Lastly, along with Daniel Liu (a collaborator and recent graduate from UCLA) and Noah Cape (a student at Williams College whom Patro advised as part of the CBCB’s REU program), Patro is developing a universal adaptor tool for single-cell sequencing analysis, accommodating different data formats and protocols. This tool will enable existing single-cell expression quantification tools to process diverse types of data without requiring code modifications for each new input format.
Patro’s winning proposal was one of 32 that were funded this year through CZI’s Essential Open-Source Software for Science program, which supports open-source software projects that are essential to biomedical research. Its goal is to support software maintenance, growth, development and community engagement for these critical tools. This year’s program co-funders are the Kavli Foundation and the Wellcome Trust.
This marks the second time that Patro has received funding from CZI. In 2022, he was awarded $350K to improve upon a collection of interrelated tools his lab developed to process genomic data.
—Story by Melissa Brachfeld, UMIACS communications group
Cummings Employs Machine Learning to Tackle Addiction Crisis

Baltimore is among the epicenters of the nationwide substance abuse crisis. The city was recently named the “Overdose capital of the U.S” after a joint investigation by The New York Times and Baltimore Banner found it averaged 1,000 overdose deaths per year, a death rate nearly double of any other large American city.
In a concerted effort to curtail overdose deaths and improve treatment for people struggling with substance abuse, researchers at the University of Maryland (UMD) and University of Maryland, Baltimore (UMB) have partnered with a regional drug treatment center to apply machine-learning techniques to electronic medical record (EMR) data.
By using EMR to analyze data from patients including race, gender, location, traumas suffered, educational background and income, the cross-institutional team is hoping to identify trends that allow treatment to be better tailored to the individual and improve recovery rates.
“Our complex histories, individual abilities and family situations are all very personal to us, but there's going to be some commonalities among people suffering from addiction,” says Michael Cummings, a professor of biology with an appointment in the University of Maryland Institute for Advanced Computer Studies (UMIACS) who is one of the researchers on the project.
Cummings’ partners on the project include Edward Bernat, an associate professor of psychology at UMD, and Fadia Shaya, a Distinguished University Professor in the school of pharmacy at UMB. Yi Chen, a fifth-year doctoral student in UMD’s biological sciences program advised by Cummings, is also involved with the project.
Their work is supported by a $50,000 grant from the Institute for Clinical and Translational Research and the University of Maryland, Baltimore.
Bernat has extensively researched substance use disorders and its intersection with psychopathology. For the past few years, he has been working directly with Tuerk House— an inpatient drug center and crisis stabilization center in West Baltimore that sees 500–600 patients a month. Bernat provides ongoing clinical intake diagnostic services for the center’s clinical team.
Not long after he began conducting evaluations of the EMRs for patients arriving at Tuerk House, Bernat saw a need to involve machine learning to get the most out of the data he was viewing. This was due to the larger amount of data, from a larger number of patients.
“That’s where machine learning often has a big advantage—going through all of those different variables, and getting it crunched down into what’s actually meaningful with the minimal data available,” says Bernat.
The researchers plan to run machine learning algorithms on up to 10,000 of Tuerk House’s unique intake records from 2022 and 2023, with a focus on additional hurdles that Black substance users face that can impact their path to recovery.
African Americans complete substance abuse treatment only 40% of the time, which is 10% less than their White counterparts, according to a 2016 study published in the Journal of Substance Abuse Treatment. The study said that factors like socioeconomic status— specifically high school attainment and employment—are both associated with higher completion rates.
“If we find some subpopulations of people with certain characteristics are more at risk for relapse or something like that, then we can modify treatment for those sets of individuals.,” Cummings says.
Tuerk House’s partnership with UMD researchers for this project is part of a broader developing collaborative relationship to advance health disparities and treatment research relevant to Black substance users.
The inpatient drug center is in a majority black city that has been disproportionately damaged by the addiction epidemic, one that Chen is incredibly familiar with.
Chen moved to Baltimore to attend Johns Hopkins University, where he completed his B.A. in biology in 2014. For four years he did community service at Stepping Stone Ministry, a faith-based organization on the Hopkins campus that gave aid to the local homeless community, only stopping in 2020, when COVID–19 forced him to do so.
“Substance use disorder and homelessness are highly comorbid,” Chen says. “So, in some ways, this project is another way of serving the same population from that part of my life using a different set of skills.”
This project contrasts Chen’s previous work which has centered around molecular biological data sets, posing occasional challenges in refining and understanding data.
Chen noted the significance of the collaboration of experts from across various fields for this project.
“Substance use disorder is a complex disorder,” he says. “This project can hopefully address that by virtue of being a collaborative work involving psychopathology, social determinants, and machine learning.”
Looking ahead, Bernat envisions a dashboard wherein a patient completes the intake assessment at Tuerk House, and the system the research team is building will suggest individualized treatment plans based on the data received.
“Our local goal with is to develop something with [Tuerk House] clinicians that they can use quickly and efficiently,” he says.
Bernat and Cummings both emphasized that the overall goal is to understand different populations and improve the success rate of addiction treatment through a data-driven, personalized approach.
“That’s the ultimate benchmark we’re looking to achieve,” says Cummings. “Hopefully, data science and machine learning can provide some insights on how specialized health care can be improved and become more individualistic.”
— Story By Shaun Chornobroff, UMIACS communications group
Using Machine Learning to Develop a More Effective Malaria Vaccine
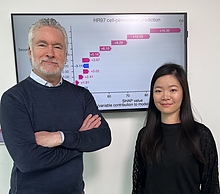
Caption: Michael Cummings (left) and Renee Ti Chou (right) collaborated on the project with researchers from the University of Maryland School of Medicine.
While cases of malaria have been decreasing in some parts of the world thanks to drug-based therapies, efforts to eradicate this life-threatening disease have recently stalled, due in part to the parasites that transmit malaria becoming resistant to current therapeutics.
Computational biologists from the University of Maryland have partnered with researchers at the University of Maryland School of Medicine to address this challenge. Using a novel approach called reverse vaccinology, which employs powerful bioinformatic tools and reverse pharmacology practices, the researchers are examining the genetic makeup of several parasites that cause malaria, seeking specific antigens to target with new vaccines.
The team’s initial research was just published in npj Systems Biology and Applications, a leading journal that focuses on scientific work that takes a systems-based approach.
The paper’s lead author is Renee Ti Chou, a data scientist at Lexical Intelligence in Rockville, Maryland, who earned her Ph.D. in computational biology last year at the University of Maryland.
Co-authors include Michael Cummings, a professor of biology with an appointment in the University of Maryland Institute for Advanced Computer Studies, and Amed Ouattara, Matthew Adams, Andrea A. Berry and Shannon Takala Harrison, all from the University of Maryland School of Medicine’s Center for Vaccine Development and Global Health.
Without a good vaccine, the researchers say, it is hard to get rid of any disease, malaria included. But to make a truly effective vaccine for malaria—which killed an estimated 608,000 people in 85 countries in 2022—scientists need to target different life cycle stages of the P. falciparum parasite, the deadliest species of Plasmodium that causes malaria in humans.
This can be tricky, though, because not only does the parasite take on different forms during its life cycle, but its genetic proteins also change, making it difficult for the human immune system to counter. So far, most vaccine efforts have focused on a few proteins without looking at the whole picture of the P. falciparum parasite’s genes.
With the reverse vaccinology approach, based in-part on powerful machine learning algorithms, the UMD/UMB researchers hope to gain a clearer picture that will lead to better vaccine antigens.
For their work, the researchers analyzed thousands of proteins from the P. falciparum parasite, considering 272 different factors for each. They used a machine learning technique called positive-unlabeled learning to sort through this data, focusing on proteins that resemble known vaccine targets.
They do not have to start with specific criteria; instead, they let the computer learn from what is already known about effective vaccine targets. This method looks at all the genes of the malaria parasite to find ones that might make good vaccine targets, even if they are not obvious at first.
Cummings, who is also the director of the Center for Bioinformatics and Computational Biology, says this approach has not been widely used for malaria, but it could help find new vaccine candidates, especially for parts of the P. falciparum parasite that are not well understood yet.
The researchers not only identified new potential vaccine targets, Cummings adds, but also ranked them based on importance. To prioritize the most promising candidates, they looked at factors like gene essentiality and when the proteins are active in the parasite’s life cycle.
“These findings offer a flexible framework for future vaccine research,” says Cummings, who was Chou’s academic adviser at UMD. “We can adjust our criteria and even apply this approach to other diseases beyond malaria. It’s a big step forward in the quest for better vaccines.”
The team’s research is supported by the National Science Foundation, the National Institutes of Health (NIH), a National Health and Medical Research Council grant, and a grant from MPowering Maryland. Other support came in the form of the Anne G. Wylie Dissertation Fellowship, which Chou received in her final year of Ph.D. training at UMD.
For her doctoral work related to this project, Chou recently received the Charles A. Caramello Distinguished Dissertation Award. Also, in work that is peripherally tied to this current research, Cummings was the recipient of an NIH grant in 2023 to study the body’s immune response to malaria to help scientists develop more effective vaccines.
—Story by Melissa Brachfeld, UMIACS communications group
The New Frontier of Microbiome Science: Computational Challenges and Solutions
The microbiome refers to the whole sum of microorganisms in a particular environment, such as the collective sum of gut bacteria in a human being. Microbiome research is a new frontier of scientific exploration. Studies that use big data technology to examine whole genomes of hundreds of organisms simultaneously represent a field called metagenomics. As this field matures, scientists are increasingly recognizing the need for sophisticated tools and technologies to decipher the complexities hidden within these microbial ecosystems.
To that end, on April 2, Mihai Pop, a professor in the Department of Computer Science and the director of the Institute for Advanced Computer Studies at the University of Maryland, gave a talk on the analytical challenges of microbiome science and how they can be combated by computational methods. The talk focused on the pivotal role of computational tools in unraveling the secrets of microbiomes and addressing the challenges associated with analyzing the vast datasets generated by these studies.
A key focus of metagenomics is the taxonomic classification of different microbes. The primary method for organizing and classifying microbes is comparing them to a database of known organism sequences. These similarity-based techniques are especially effective when the organisms in the sample are well represented in the database. Pop mentioned one of the most common similarity search methods used to classify microorganisms, the Basic Local Alignment Search Tool (BLAST). However, BLAST often misidentifies the closest neighbor to the microorganism of interest; the “most similar” organism according to BLAST may not actually be the most closely related.
“How can we find what’s the real [closest hit] if there is a hit? The E-value is misleading,” Pop explained during the talk, suggesting that BLAST may not always accurately identify the most similar organism to the microbiome of interest.
The E-values Pop mentioned refer to parameters in BLAST that describe the number of hits one can “expect” to see by chance when searching a database of a particular size. Pop also emphasized how many of these problems were only discovered years after BLAST integrated into common use.
“These are things we found out 20, 30, 40 years after [the computational tool] was written ... even though something has been used for many, many years, there [are] still things to learn about it,” Pop explained.
One of the other main challenges Pop highlighted is how the structure of biological databases affects scientists’ ability to reliably reveal insights on the microbiome. Reference databases are not all-encompassing. Many microorganisms cannot be cultured in labs, and a large proportion of those that can have not been sequenced or added to these reference databases. Consequently, not all environmental organisms are included in the sequence database, which limits the accuracy of similarity-based methods.
These problems are further compounded by the lack of contiguous information available in most sequencing datasets. Many sequencing analyses have to begin by joining together many sequence fragments and stitching together a whole related sequence. Assembling the sequencing data is also an unstandardized process, as new technologies used for assembling genomes are constantly being developed. These limitations can impede researchers' ability to derive meaningful insights and connections from microbiome datasets, because it substantially limits precision and decreases the accuracy of reference databases.
Pop then transitioned to discussing algorithms and software approaches to sequence similarity. Many current software used in classification employ the most recent common ancestor (MRCA) method. MRCA provides an annotation (marking of a specific feature of the DNA sequence) at the broadest taxonomic class that encompasses all of the possible markings in a sequence. However, this means that different types of software that use MRCA only make a few classifications at the genus or species level, meaning that stronger relationships between two microbes cannot be determined at the family, class or phylum level.
To address this challenge, Pop shared efforts from his own lab to develop advanced computational tools tailored specifically for microbiome analyses. He specifically focused on the Ambiguous Taxonomy eLucidation by Apportionment of Sequences (ATLAS). ATLAS is a data-driven database partitioning method, which aims to divide a large dataset into smaller, more easily analyzable datasets. ATLAS groups sequences into biologically meaningful partitions by querying the sequence against a reference database and then identifying and clustering hits that are considered significant. ATLAS also represents a shift away from the MRCA method.
As the talk concluded, Pop emphasized the critical need for interdisciplinary collaboration to advance microbiome research. Integrating expertise from fields such as biology, computer science and statistics is essential for developing innovative solutions to microbiome-related challenges. This interdisciplinary approach allows researchers to harness the power of computational tools to extract meaningful patterns and associations from microbiome datasets.
—This article by Shreya Tiwari was originally published in The News-Letter, a student publication at John Hopkins University.
Marking Five Years of Research, Scholarship and Innovation in the Iribe Center

A well-designed building is more than just a structure—it’s also a catalyst for collaboration, fostering a dynamic environment where people come together to brainstorm ideas and create new technologies.
The Brendan Iribe Center for Computer Science and Engineering is exactly such a place. Launched five years ago this month, the 215,000 square foot structure is home to the Department of Computer Science and the University of Maryland Institute for Advanced Computer Studies (UMIACS).
A visit to the ground floor of Iribe offers multiple experiences. Computer science is the largest major on campus—and one of the largest in the nation—with visitors keenly aware of this fact as they observe hundreds of undergraduates heading to class or studying in the expansive lobby area.
Cutting-edge research is also visible on the ground level, with a shiny silver mannequin staring at you from the Small Artifacts Lab (it’s used to test wearable technologies), miniature drones buzzing in the Brin Family Aerial Robotics Lab, and digital art or other 3D visuals captivating your attention as you walk by the Immersive Media Design lab space.
For UMIACS, the Iribe Center is a boon for the seamless flow of bold ideas and new knowledge across multiple academic disciplines.
The institute currently supports more than 80 faculty and 200 Ph.D. students from 15 departments on the UMD campus. Most of these faculty and students reside in the Iribe Center, spread out in offices, labs and workspaces on the third and fourth floor of the building as well as part of the fifth floor that’s home to the Maryland Cybersecurity Center.
“Most scientific advances that have transformed our world have emerged at the boundary between disciplines,” says Mihai Pop, a professor of computer science who is the director of UMIACS. “Our continued scientific leadership critically depends on scientists from multiple disciplines coming together to work on impactful projects.”
For Naomi Feldman, a professor of linguistics, the Iribe Center has offered abundant opportunities to interact with faculty she might not normally see in her home department.
“I’m a computational linguist and having colleagues literally right down the hallway working on computational acoustics, computer vision and machine learning has been very advantageous for me and for my students,” Feldman says.
The building has tremendous aesthetic value as well, faculty members say.
Vanessa Frias-Martinez, an associate professor in the College of Information Studies, says the airiness of the Iribe Center and the stunning views it offers make for a calm and creative workspace. 
“My office has large windows and plenty of natural light, and we have our beautiful rooftop garden—both serve as a source of inspiration and comfort,” says Frias-Martinez, who is director of the Computational Linguistics and Information Processing (CLIP) Lab. “I feel like I can really focus on my work and be productive, but it’s also nice to pause and unwind with the fantastic views and green spaces we have available.”
Feldman, also a member of the CLIP Lab, says that the Iribe Center has multiple spaces for students to meet and network and for faculty to host workshops and seminars.
“Our students come from diverse academic backgrounds—linguistics, computer science, information studies, human-computer interaction, and more—and the workshops and talks in the Iribe Center offer a robust opportunity to communicate across disciplines,” Feldman says.
Michael Cummings, a professor of biology who leads the Center for Bioinformatics and Computational Biology, says that in addition to his graduate students having a modern, inviting workspace, moving to the Iribe Center has made many of his day-to-day administrative tasks easier and more productive.
“I’m able to walk down the hall and talk with the business office on the status of a research grant or meet with our tech staff to go over our computing needs,” Cummings says. “This is not only more efficient, but also more pleasant.”
With research and innovation in UMIACS constantly expanding, the Iribe Center will continue to serve its mission, and serve it well, says UMIACS director Pop.
“We’re thankful for the private donations and state support that made this building possible,” Pop says. “But it remains up to the people that work here—faculty, students and staff—to fulfill the vision of why this space was built: we’re a hub for technology, collaboration and discovery that will have an impact on the world.”
—Reporting on this story by Melissa Brachfeld, UMIACS communications group
UMIACS Expands Data Center to Support Research Growth
Jisha Jesudass (right in photo) is responsible for designing and maintaining a sophisticated network system used by UMIACS faculty to seamlessly move data among various stakeholders both on and off campus.
For almost four decades, a powerful data center in the A.V. Williams Building has been an essential part of the University of Maryland Institute for Advanced Computer Studies (UMIACS), supporting the institute’s mission of incentivizing cutting-edge research and staying at the forefront of technological innovation.
As the institute’s research portfolio continues to expand, the infrastructure needed to support it—particularly in compute-heavy areas like artificial intelligence (AI) and machine learning—also needs to grow.
In April, UMIACS data center staff began coordinating a three-month renovation to expand the center’s physical footprint by 50 percent, growing from 2,300 square feet to 3,450 square feet.
The expansion project will provide additional space for computing hardware and will offer better options to ensure that the data center’s power grid and cooling systems can handle any increased workload, says Derek Yarnell, the institute’s director of computing facilities.
Much of the current workload is handled by multiple racks of graphical processing units (GPUs), powerful parallel computing hardware used for gaming, content creation—and in the case of UMIACS—machine learning and neural networks for high performance computing in areas like AI and computer vision.
In the past five years, UMIACS has increased the number of GPUs in its data center by almost tenfold, Yarnell says. This has allowed researchers to perform certain tasks at almost one petaFLOP of speed—equivalent to more than one quadrillion floating-point operations per second.
“If you measure the amount of computational infrastructure in place that is dedicated specifically toward AI and machine learning research, we are the largest on campus by a wide margin,” Yarnell says. “And we believe this is only the beginning.”
UMIACS faculty are constantly seeking new mechanisms to upgrade their computing resources, says Tom Goldstein, a professor of computer science and director of the University of Maryland Center for Machine Learning (CML).
Goldstein advocates for making economy of scale purchases, wherein CML faculty collectively pool portions of their individual research grants to purchase racks, routers, wiring and accessories needed to support multiple GPU clusters.
The data center expansion matches well with that concept.
“We want to be forward-looking in regard to increasing our computing capabilities, particularly those of us requiring high-memory nodes for projects that focus on large language models and other emerging AI research,” Goldstein says.
Other UMIACS faculty also want to improve both the availability and level of computing power available through the institute’s data center.
Furong Huang, an assistant professor of computer science with an appointment in UMIACS, is working with her graduate students to build a foundation model for sequential decision making, a form of generalist AI that holds the capacity to learn and master diverse sets of tasks downstream.
“Imagine an AI that not only learns quickly, but adapts to your daily tasks autonomously,” says Huang, who is a member of CML.
This work requires analyzing massive amounts of data used in the AI system, a task best handled by a large network of multiple GPUs, Huang says.
Supporting these myriad research efforts falls on Yarnell’s team, a dedicated staff of network engineers and high-performance computing experts assisted by more than 20 undergraduates working at the UMIACS Help Desk.
The institute’s technical staff are involved in the computing process from start to finish, Yarnell says. This includes helping faculty spec out their equipment needs to submit in their research proposals; working with UMD procurement to purchase the equipment; installing all the hardware and software when it arrives on campus; and maintaining the equipment over its technical lifespan by providing security patches and updates.
The tech staff is also responsible for managing and securely storing all the data used in UMIACS research. Current storage capabilities top out at around 3.65 petabytes, with that number constantly increasing, says UMIACS network engineer Jisha Jesudass.
There has also been steady growth in the network capabilities for the institute. When UMIACS moved from the A.V. Williams Building to the Brendan Iribe Center for Computer Science and Engineering in 2019, Jesudass’ job responsibilities included designing and maintaining a high-capacity network—miles of wiring and dozens of sophisticated routers—used to transmit large volumes of data both on and off campus.
She emphasized that the upgrades currently underway in the data center include further improving connectivity capabilities to make the workflow of UMIACS projects more efficient.
“Every job or every researcher right now, they want to minimize latency and delays. This new infrastructure will help a lot with that,” Jesudass says.
For Yarnell (pictured left), the renovation and expansion of the data center remains a work in progress and a labor of love. His involvement with the data center started in 1998 as an undergraduate working at the UMIACS Help Desk. Since that time, he’s seen—and helped lead—much of the data center’s growth.
“Our data center has obviously stood the test of time up to this point—it’s like a Ferrari, a fine-tuned machine that has aged very well,” he says. “But we still think we can continue to innovate it, and continue to build it, in a way that will allow the center to support any new opportunities that are yet to come.”
—Reporting on this story by Shaun Chornobroff, UMIACS communications group
University of Maryland Hosts Microbiome Research Symposium

More than 80 people braved stark wintry conditions on January 16 to attend a research symposium at the University of Maryland that explored the world of complex microbial communities.
The Symposium on Microbiome Research at the Interface of Environment, Health and Agriculture joined researchers from the federal government, academia and private industry who are focused on the connectivity between microbes interacting with each other, the environment, agricultural systems, and human and animal health.
Hosted by the University of Maryland Center of Excellence in Microbiome Sciences, the event featured multiple talks, breakout sessions, an engaging poster session, and a networking reception. All the events went off without a hitch, despite 4 inches of snow that closed the university for the day and made travel difficult.
“We were fortunate that more than two-thirds of the people who registered were able to show up and participate,” says Mihai Pop, a UMD professor of computer science who is the director of the microbiome center. “We were particularly pleased by the strong turnout from federal scientists in the region, as well as colleagues from the medical and dentistry schools in Baltimore.”
A morning keynote talk by Susannah Tringe, division director of the DOE Joint Genome Institute at the Lawrence Berkeley National Laboratory, looked at the sequence-based interrogation of soil microbiomes, and how those microbes can benefit various ecosystems.
The afternoon keynote by Joff Silberg, a professor of biosciences at Rice University, was presented virtually as Silberg was unable to fly out of Houston due to poor weather. His talk explored the use of engineered living microbes, and how they might be used to monitor various soil pollutants in real time.
Other talks included how microbial communities can impact coffee growers, the effect of cow manure microbes on farm soil, microbial activity related to women’s gynecologic health, and other topics focused on human gut bacteria and inflammatory bowel disease.
“There’s such a rich diversity of perspectives and ongoing work at the University of Maryland involving microbiome sciences,” says Hannah Zierden, an assistant professor of chemical and biomolecular engineering at UMD and core member of the microbiome center. “I’m excited at the opportunities we have and look forward to continued collaborations—as well as new ones—as we expand our outreach and impact.”
Zierden presented some recent research from her own UMD lab at the conference, which aims to better understand the function of bacterial extracellular vesicles produced by vaginal microbes, and how they might be used to engineer biocompatible therapies for healthy pregnancies.
A large contingent of researchers from the University of Maryland School of Dentistry were onsite for the symposium, including Areej Alfaifi, a doctoral student in the dental biomedical sciences program.
“This event broadened my perspective by introducing me to entirely different aspects of microbiome studies,” says Alfaifi, whose dissertation explores the use of genomic sequencing tools to gain a deeper understanding of the oral microbiome in COVID-19 patients. “Connecting with students and faculty from different schools was an amazing experience that reshaped my thoughts on the field. This meeting was truly unforgettable!”
Additional attendees included faculty, postdocs and graduate students from the University of Delaware, Towson University, University of Maryland School of Medicine, and the University of Maryland, College Park.
Federal scientists in attendance hailed from the USDA, FDA, Department of Energy, and the Smithsonian National Zoo, with representatives from QIAGEN, CosmosID—both major sponsors of the symposium—also present.
The symposium also received support from the University of Maryland Institute for Advanced Computer Studies, Mid-Atlantic Microbiome Meet-up, and the UMD’s Grand Challenges Grants program.
Pop said the UMD microbiome center will help coordinate another symposium in 2025 in Baltimore, working closely with the Institute for Genome Sciences at the University of Maryland, Baltimore to investigate new topics related to microbiome sciences.
“We expect to continue our momentum in this area, which reaches across multiple scientific, medical and policy-related disciplines,” Pop says. “Our belief is that the basic unresolved questions involving microbial communities are interrelated—and so are the solutions we’re working on.”
—Story by Maria Herd, UMIACS communications group
What Makes Urine Yellow? UMD Scientists Discover the Enzyme Responsible
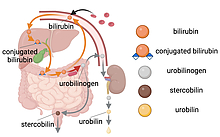
Researchers at the University of Maryland and National Institutes of Health have identified the microbial enzyme responsible for giving urine its yellow hue, according to a new study published in the journal Nature Microbiology on January 3, 2024.
The discovery of this enzyme, called bilirubin reductase, paves the way for further research into the gut microbiome’s role in ailments like jaundice and inflammatory bowel disease.
“This enzyme discovery finally unravels the mystery behind urine’s yellow color,” said the study’s lead author Brantley Hall, an assistant professor in the University of Maryland’s Department of Cell Biology and Molecular Genetics. “It’s remarkable that an everyday biological phenomenon went unexplained for so long, and our team is excited to be able to explain it.”
When red blood cells degrade after their six-month lifespan, a bright orange pigment called bilirubin is produced as a byproduct. Bilirubin is typically secreted into the gut, where it is destined for excretion but can also be partially reabsorbed. Excess reabsorption can lead to a buildup of bilirubin in the blood and can cause jaundice—a condition that leads to the yellowing of the skin and eyes. Once in the gut, the resident flora can convert bilirubin into other molecules.
“Gut microbes encode the enzyme bilirubin reductase that converts bilirubin into a colorless byproduct called urobilinogen,” explained Hall, who has a joint appointment in the University of Maryland Institute for Advanced Computer Studies and is a core faculty member in the Center for Bioinformatics and Computational Biology. “Urobilinogen then spontaneously degrades into a molecule called urobilin, which is responsible for the yellow color we are all familiar with.”
Urobilin has long been linked to urine’s yellow hue, but the research team’s discovery of the enzyme responsible answers a question that has eluded scientists for over a century.
Aside from solving a scientific mystery, these findings could have important health implications. The research team found that bilirubin reductase is present in almost all healthy adults but is often missing from newborns and individuals with inflammatory bowel disease. They hypothesize that the absence of bilirubin reductase may contribute to infant jaundice and the formation of pigmented gallstones.
“Now that we’ve identified this enzyme, we can start investigating how the bacteria in our gut impact circulating bilirubin levels and related health conditions like jaundice,” said study co-author and NIH Investigator Xiaofang Jiang. “This discovery lays the foundation for understanding the gut-liver axis.”
In addition to jaundice and inflammatory bowel disease, the gut microbiome has been linked to various diseases and conditions, from allergies to arthritis to psoriasis. This latest discovery brings researchers closer to achieving a holistic understanding of the gut microbiome’s role in human health.
“The multidisciplinary approach we were able to implement—thanks to the collaboration between our labs—was key to solving the physiological puzzle of why our urine appears yellow,” Hall said. “It’s the culmination of many years of work by our team and highlights yet another reason why our gut microbiome is so vital to human health.”
###
This article was adapted from text provided by Brantley Hall and Sophia Levy.
In addition to Hall, UMD-affiliated co-authors included Stephenie Abeysinghe (B.S. ’23, public health science); Domenick Braccia (Ph.D. ’22, biological sciences); biological sciences major Maggie Grant; biochemistry Ph.D. student Conor Jenkins; biological sciences Ph.D. students Gabriela Arp (B.S. ’19, public health science; B.A. ’19, Spanish language), Madison Jermain, Sophia Levy (B.S. ’19, chemical engineering; B.S. ’19, biological sciences) and Chih Hao Wu (B.S. ’21, biological sciences); Glory Minabou Ndjite (B.S. ’22, public health science); and Ashley Weiss (B.S. ’22, biological sciences).
Their paper, “Discovery of a gut microbial enzyme that reduces bilirubin to urobilinogen,” was published in the journal Nature Microbiology on January 3, 2024.
This research was supported by the NIH’s Intramural Research Program, the National Library of Medicine and startup funding from UMD. This article does not necessarily reflect the views of these organizations.
Colwell Honored with Builders of Science Award from Research!America

A noted University of Maryland researcher is being honored for their pioneering insights into microbial water quality surveillance and longstanding efforts in fighting waterborne diseases on a global scale.
Distinguished University Professor Rita Colwell was recently named a recipient of the 2024 Builders of Science Award, one of the top Advocacy Awards given each year by Research!America, a nonprofit alliance that supports increased funding and better policies related to medical and health research.
The Builders of Science Award is specific to those who have provided leadership and determination in building an outstanding scientific research organization, as well as those who have been at the forefront of scientific research.
Still active in research through her appointment in the University of Maryland Institute for Advanced Computer Studies (UMIACS), much of Colwell’s scientific career has been focused on tracking and predicting outbreaks of dangerous waterborne pathogens by combining bioinformatics with satellite imaging. She recently published a report that identified several dangerous bacteria in Florida’s coastal waters following Hurricane Ian in 2022.
The impact of Colwell's research and scholarship has been especially felt in cholera-endemic countries worldwide—the result of a key discovery she made in the 1970s involving cholera-causing bacteria, known as Vibrio cholera. Previously thought to be incapable of surviving more than a few hours outside a human host, she discovered that the bacteria occur naturally in the aquatic environment associated with plankton.
This highlighted the critical link between the environment and cholera and led to Colwell's subsequent application of satellite imagery and modelling to predict cholera outbreaks, as well as her innovative use of sari cloths as filters to greatly reduce contamination in drinking water.
Cowell was the 11th director and first woman to lead the National Science Foundation, and during her tenure (1998–2004), she oversaw its most significant period of growth. She also championed and secured NSF funding for innovative science and engineering education programs and initiatives to advance women in academic engineering and science careers.
Other notable awards and recognition that Colwell has received include the National Medal of Science; the Stockholm Water Prize; membership into the National Academy of Sciences, Royal Society of Canada, Swedish Royal Academy of Science, Irish Royal Academy of Science, and the Bangladesh and Indian academies of science; and the “The Order of the Rising Sun, Gold and Silver Star” from the emperor of Japan.
Colwell and the other Advocacy Award recipients will be officially recognized on March 13, 2024 at an event at the National Academy of Sciences in Washington, D.C.
New Study Confirms Presence of Dangerous Bacteria in Florida’s Coastal Waters Following Hurricane Ian

Caption: Hurricane Ian is pictured from the International Space Station as it orbited 258 miles above the Caribbean Sea on September 26, 2022. At the time of this photograph, Ian was just south of Cuba gaining strength and heading toward Florida.
When Hurricane Ian struck southwest Florida in September 2022, it unleashed a variety of Vibrio bacteria that can cause illness and death in humans, according to a new study published in the journal mBio.
Using a combination of genome sequencing and satellite and environmental data, a team of researchers from the University of Maryland, the University of Florida and microbiome company EzBiome detected several pathogenic Vibrio species in water and oyster samples from Florida’s Lee County, a coastal region that was devastated by Hurricane Ian. The samples, which were collected in October 2022, revealed the presence of two particularly concerning species: Vibrio parahaemolyticus and Vibrio vulnificus.
“We were very surprised to be able to detect—without any difficulty—the presence of these pathogens,” said the study’s senior author Rita Colwell, a Distinguished University Professor in the University of Maryland Institute for Advanced Computer Studies (UMIACS) who has studied Vibrio for the last 50 years.
The study’s findings correspond with a reported increase in V. vulnificus cases in the state of Florida in October 2022. According to the Florida Department of Health, Lee County—which had the highest caseload in the state—reported 38 infections and 11 deaths linked to vibriosis.
Vibrio bacteria naturally occur in the ocean, where they live symbiotically with crustaceans, zooplankton and bivalves. When the bacteria encounter humans, some species can cause an infection known as vibriosis, but the side effects depend on the type of Vibrio and severity of the infection. V. parahaemolyticus can cause gastroenteritis and wound infections, while the V. vulnificus species can cause necrotizing fasciitis—a flesh-eating infection—and kills 1 in 5 infected people.
People can contract vibriosis by eating raw or undercooked seafood or by getting seawater in an open wound. Because Vibrio thrive in warm saltwater, hurricanes and floods can increase the chances of a person becoming exposed.
Several conditions during and after Hurricane Ian favored the growth of Vibrio bacteria, including the amount of rainfall, changes in sea surface temperature and concentrations of chlorophyll in the ocean, which can indicate densities of phytoplankton—and subsequently zooplankton—in an area. In places with plankton blooms, the researchers found an abundance of Vibrio bacteria.
With warming oceans expected to fuel wetter and more powerful storms like Ian, coastal communities could see more Vibrio infections in the future.
“These Vibrios generally grow well between 15 and 40 degrees Celsius [59–104 degrees Fahrenheit], so as the temperature warms, their generation time shortens and they divide faster and faster,” Colwell said. “The warming of seawater—which mixes with freshwater, creating optimal salinities—really enhances the growth of Vibrios, so it’s a very serious concern.”
While the environmental conditions in Florida following Hurricane Ian were ripe for vibriosis, these cases are not limited to southern climes. In August 2023, three people in New York and Connecticut died from V. vulnificus infections.
Colwell and her co-authors—which included Kyle Brumfield (Ph.D. ’23, marine estuarine environmental sciences) and UMD Cell Biology and Molecular Genetics Research Professor Anwar Huq—predicted this recent spike in vibriosis cases based on trending environmental conditions in the Northeast United States. As ocean temperatures continue to rise, Colwell said the rapidly warming Chesapeake Bay could also be affected.
“The waters are much warmer in Florida right now than they are in the Chesapeake Bay, but on a lot of the East Coast, the waters are warming,” Colwell said. “This is a threatening indication that we may be seeing more Vibrio vulnificus infections.
Colwell and her co-authors noted that while they analyzed only a limited number of samples, their findings illustrate the potential of genetic analysis, environmental data and remote sensing to improve public health by proactively detecting and characterizing Vibrio pathogens.
They also called for further investigation to quantify the prevalence of Vibrio bacteria in different locations, seasons and environmental conditions. Colwell said this research is not only vital to public health but also an important step in understanding our changing climate.
“On the positive side, knowing that these infections are associated with the increased variability of a changing climate, perhaps now is the time to develop mechanisms to understand and mitigate it,” Colwell said. “Climate change and flooding are clearly linked to infectious disease, and we need to take it seriously.”
###
The paper “Genomic Diversity of Vibrio spp. and Metagenomic Analysis of Pathogens in Florida Gulf Coastal Waters Following Hurricane Ian” was published in mBio on October 16, 2023.
—Story by Emily Nunez, College of Computer, Mathematical, and Natural Sciences communications team
