All News
Tracking Transgenic Crop-Produced Mutations in the Corn Earworm

Environmentally friendly, sustainable pesticides are popular with consumers, who are more likely to purchase sweet corn from their local market that hasn’t been sprayed with potentially harmful chemicals.
Now, supported by a $650,000 grant from the U.S. Department of Agriculture (USDA), two University of Maryland researchers have teamed up to help ensure these pest-fighting methods remain on the table, in part by understanding why a common pest known as the corn earworm, or H. zea, has developed resistance to pesticides widely used in farming.
The researchers—Megan Fritz, an associate professor of entomology with an affiliate appointment in the University of Maryland Institute of Advanced Computer Studies (UMIACS), and Erin Molloy, an assistant professor of computer science who is also in UMIACS—plan to share the knowledge with federal regulators, private industry and agricultural producers, providing strategies to monitor for and manage growing pesticide resistance before a crisis arises.
They’re particularly interested in tracking genetic mutations that occur in insects that ingest proteins from Bacillus thuringiensis, or Bt, a bacterium that occurs naturally in soil and has been incorporated into some types of corn. These proteins are not toxic to humans, but have proven effective against agricultural pests like the earworm.
Bt-corn is a genetically modified, or transgenic crop that produces the proteins as it grows, and is widely used for cornmeal, tortilla chips, and other human and animal food products. Fritz and others previously showed that over time, H. zea that ingested Bt crops developed resistance to the Bt proteins expressed by transgenes in the plant. Now the researchers want to determine whether signals of resistance can be detected early—before resistance scales up to a level that presents significant challenges to farmers.
The earworm is practically everywhere, and mutations spread quickly among populations, said Fritz, the project’s principal investigator. Eating transgenic corn with just enough Bt protein to impact H. zea’s growth and development, but not kill all corn-feeding caterpillars may be responsible. This “low dose” of Bt protein allowed mutations that likely existed in the H. zea genome before these crops were widely used to accumulate over time.
The UMD researchers are trying to identify when the mutations arose and their frequency across North America by tracking H. zea’s genome to identify signals of resistance evolution to environmentally friendly pesticides.
They believe their work might determine whether other pest species could benefit from similar genomic monitoring.
“Many of the pest species that are most problematic are like H. zea: They have similar genetic and population demographic qualities that might make them amenable to genomic resistance monitoring,” says Fritz, who is an affiliate member of the Center for Bioinformatics and Computational Biology (CBCB).
Molloy is handling much of the computational end of the project, with help from first-year computer science Ph.D. student Junyan Dai.
“There are currently no computational tools for detecting certain mutations commonly involved in resistance evolution from genomic monitoring datasets collected over time," says Molloy, who is also a member of CBCB. "The goal of our interdisciplinary USDA project is to bridge this gap."Also assisting on the project is Kate Taylor, a former postdoctoral researcher in Fritz’s lab who is now an assistant professor of computational biology at Hofstra University. Fritz credits Taylor with much of the foundational research leading up to the current USDA project.
Another team member is Abuzar Bhatty, a third-year Ph.D. student in UMD’s Computational Biology, Bioinformatics and Genomics Program. Bhatty is developing whole genome simulations of H. zea and other insect species that may provide answers for scientists researching other pest species.
This includes the notorious Colorado potato beetle, whose historical levels of resistance to multiple pesticides left farmers stranded with few pest management options; at some points in history, flamethrowers became the most viable option for their control.
Ultimately, Fritz’s hope is that this interdisciplinary project that joins entomologists with computer scientists will help prevent the loss of these transgenic pesticides to a “tragedy of the commons” situation.“Susceptibility to an insecticide, or a pesticide, is one of those common goods no one thinks about—everybody loves [sustainable pest management] until it’s gone," she says. "And when it’s gone, it impacts all of us.”
—Story by Kathryn Klett, UMIACS communications group
CBCB Team Publishes Innovative Research on Improving Parkinson’s Disease Diagnosis
Researchers in the University of Maryland’s Center for Bioinformatics and Computational Biology (CBCB) have just published research that may soon help doctors better diagnosis patients with Parkinson’s disease.
Their work, done in tandem with researchers at the University of Maryland, Baltimore and others, uses machine learning algorithms to analyze data from wearable sensors that track movement. Ultimately, the researchers say, this can lead to more accurate and earlier diagnoses of people suffering from Parkinson’s, which in turn can lead to earlier therapeutic interventions.
Rana Khalil, a sixth-year Ph.D. student in computer science at UMD and lead author of the paper detailing the research, worked on the project with her adviser, Michael Cummings, a professor of biology who is the director of CBCB.
The project was funded by a $150,000 seed grant from the University of Maryland Strategic Partnership: MPowering the State, known as MPower, an initiative that fosters collaboration between the University of Maryland, College Park, and the University of Maryland, Baltimore.
Parkinson’s disease is a nervous system disorder that affects movement. It has a slow onset, starting with mild symptoms, such as a subtle tremor in one hand, that may progress to severe muscle rigidity and an inability to walk without help. According to the National Institutes of Health, approximately 500,000 people in the U.S. have been diagnosed with Parkinson’s.
But because many people are undiagnosed—or have been misdiagnosed—experts believe that as many as one million Americans may have the debilitating disease.
This considerable discrepancy highlights one of the central problems faced by clinicians and patients: the challenge of accurately diagnosing the disease.
“The diagnosis of mobility disorders is very difficult,” explains Cummings. “Much of the process is subjective, and as such, isn’t highly accurate.”
As currently practiced, doctors diagnose patients with Parkinson’s by asking them to do a variety of mobility tasks, observing their walking and movement patterns, and testing their reflexes. However, this process is time-consuming and requires a lot of effort from both the clinician and the patient.
“We want to use objective measures to make the process more accurate and efficient,” says Cummings, who has an appointment in the University of Maryland Institute for Advanced Computer Studies. “By improving the time- and cost effectiveness of the process, we intend to benefit health care practice.”
If successful, he adds, this improvement would alleviate the physical and psychological distress patients face by cutting down on multiple clinic visits and misdiagnoses.
Although wearable sensors to diagnose Parkinson’s have been developed in the past, their complexity has prevented doctors from using them in clinics. The new research, published in MDPI’s journal Sensors, simplifies the use of sensors and machine learning in a clinical setting and makes it more practical, Cummings says. It demonstrated that a single sensor placed on the lower back and a single mobility task involving multiple movements can effectively distinguish individuals with Parkinson's disease from controls. The researchers then developed a sophisticated machine learning framework that analyzed patterns and variations in the data.
This enhanced the identification of disease symptoms and diagnostic accuracy of the disease, with the study reporting an accuracy of 92.6% in identifying participants across different Parkinson’s stages, surpassing the accuracy of 81% previously reported for clinical diagnosis by movement disorder experts.
This significant success was preceded by numerous difficulties, such as in data collection and signals processing. However, both Khalil and Cummings acknowledged that the computing resources from the CBCB and the financial support from the MPower initiative proved essential in overcoming these challenges.
“The MPower grant provided a reliable source of support for Rana,” Cummings says. “It enabled us to work on this project, gave us structure and a clear focus, and facilitated collaboration between our team and our partners.”
Moving forward, CBCB researchers are already deeply involved in a related study on distinguishing Parkinson’s from other movement disorders, attempting to further enhance accuracy and deter misdiagnosis.
—Story by Aleena Haroon, UMIACS communications group
Hall Receives $3.5M in Federal Funding for Innovative Gut Microbiome Research

A University of Maryland expert in computational biology has been awarded significant federal funding to uncover the mysteries of the human gut microbiome.
Brantley Hall, an assistant professor of cell biology and molecular genetics with an appointment in the University of Maryland Institute for Advanced Computer Studies (UMIACS), is the recipient of two awards from the National Institutes of Health (NIH) totaling $3.5 million.
The federal funding—a $1.9 million R35 investigators award from the National Institute of General Medical Sciences and a $1.6 million R33 phased innovation grant from NIH—will support Hall’s groundbreaking work in the characterization of microbial enzymes and the development of novel wearable devices for monitoring gut health.
The gut microbiome is an intricate ecosystem composed of trillions of microorganisms, including bacteria, fungi, parasites and viruses. Each person has their own unique microbiome, which is influenced by their diet and environment. This complexity makes studying gut health considerably challenging for researchers, resulting in a lack of in-depth knowledge about the gut.
With support from the R35 award over the next five years, Hall and his research team aim to address these challenges, in part by systematically identifying and characterizing a class of microbial enzymes called ene-reductases. These enzymes play a pivotal role in the intricate interactions between gut microbes and their human hosts.
“By discovering key gut microbial enzymes, we aim to uncover how gut microbial biotransformations influence overall human health,” Hall says.
The R33 grant will provide three years of funding for Hall’s team to develop wearable devices that will enable the real-time monitoring of gut microbial activity. This project aims to translate laboratory findings into practical applications, Hall says, and has the potential to revolutionize the way gut health is measured and managed.
In a broader sense, Hall says this latest round of research funding supports work that will help his team and other scientists highlight the symbiotic relationship between humans and gut microbes. As hosts, we provide our gut microorganisms with shelter and food, and in return, they help us keep our body functioning well, he explains.
These same microorganisms break down dietary fibers and carbohydrates that we can’t digest on our own. They not only affect our digestion, but also interact with a range of organs and systems in our body.
For instance, the gut microbiome affects liver function through the gut-liver axis, a key focus of Hall's research. A recent study by Hall discovered that our gut microbes’ interactions with liver enzymes is what produces the distinct yellow color of urine.
While most gut microbes are beneficial, some bacteria can contribute to various health issues. Disorders linked to harmful microbial activity include Inflammatory Bowel Disease (IBD), which affects millions globally. Certain gut bacteria can even produce compounds that elevate risk factors for heart disease, highlighting the broad impact of the microbiome on overall health.
Hall’s research offers significant insights into understanding gut-related disorders and advancing effective treatments. However, the journey toward these breakthroughs has been marked by considerable challenges.
“Gut microbes are really hard to grow,” Hall explains. Although his team uses an anerobic chamber, an environment in which conditions like temperature and oxygen can be carefully controlled, cultivating these enzymes is an incredibly difficult task.
“Identifying the right enzyme among millions of genes is like searching for a needle in a genomic haystack,” Hall says. He and his team use a complex mix of bioinformatics and biochemistry to identify which enzymes are involved in what reactions.
Given the complex nature of gut microbiome research, providing institutional support to researchers is essential, Hall says.
At the University of Maryland Center for Excellence in Microbiome Sciences, where Hall is an active member, he works with researchers from a diverse range of disciplines, including food safety and nutrition, computer science, bioengineering, civil and environmental engineering, and more.
He also acknowledges the support from UMIACS, which provides technical and administrative services and is home to the Center for Bioinformatics and Computational Biology, which offers additional computational resources.
“I am deeply thankful to the NIH for their continued investment in microbiome research and to the centers and institutes at the University of Maryland that provide the resources and collaborative environment essential to the success of these projects,” Hall says.
—Story by Aleena Haroon, UMIACS communications group
On the Right Tract: Hall Studies the Gut Microbiome, Gastrointestinal Diseases

You could say that a gut feeling guided Brantley Hall’s career path.
After earning his Ph.D. in genetics, bioinformatics and computational biology from Virginia Tech in 2016, Hall began studying something that’s part of every person on the planet: the gut microbiome. Trillions of microorganisms form communities in the gastrointestinal tract that are vital to human health, but much is still unknown about their intricacies.
Now an assistant professor of cell biology and molecular genetics with a joint appointment in the University of Maryland Institute for Advanced Computer Studies (UMIACS), Hall works to demystify the process of digestion.
“I’m extremely happy to have chosen this field,” said Hall, who is a core faculty member in both the Center for Bioinformatics and Computational Biology and University of Maryland Center of Excellence in Microbiome Sciences.
“It’s rewarding because so many people can relate to and engage with my research. Even your choice of lunch has a huge impact on your gut microbiome.”
Hall’s lab studies the human gut microbiome and its links to medical ailments such as inflammatory bowel disease. His team developed a wearable device that measures gas produced by gut microbes—a tool that, until now, never existed. By taking these measurements in real time, Hall hopes to shed light on a range of gastrointestinal symptoms that can hamper a person’s quality of life.
Gut microbiome research has a wide range of applications in medicine. A recent study led by Hall identified the gut microbial enzyme responsible for making urine yellow.
“This enzyme discovery finally unravels the mystery behind urine’s yellow color,” Hall said. “It’s remarkable that an everyday biological phenomenon went unexplained for so long, and our team is excited to be able to explain it.”
Hall and his collaborators believe this enzyme, called bilirubin reductase, could be linked to jaundice—a condition that leads to yellowing of the skin and eyes—and plan to do a clinical trial in infants to test their hypothesis.
The gut microbiome also shares links with allergies, diabetes, arthritis, multiple sclerosis, psoriasis, Parkinson’s disease and many more conditions. Despite its importance to human health, gut microbiome research is still in its infancy. It didn’t take off until the early 2000s, with the rise of genomic sequencing and oxygen-free chambers that allowed microbes to be grown in a lab.
“Scientists have known about the importance of the gut microbiome for a long time, but there was just no one way to grow or measure the microbes,” Hall said. “We knew stuff was happening in people’s guts and that microbes were responsible, but beyond that, there wasn’t much we could do.”
While Hall’s lab grows and studies microbes in isolation, his team aims to tackle a bigger challenge: figuring out how to study the activity of microbial communities in a live human body.
“We need to measure what microbes are doing in real time—that’s the big gap in the field,” Hall said. “What we need are new tools that can measure gut microbes.”
Looking back at the last seven years, Hall is glad he followed his gut and pursued this field of research. He said gut microbiome research holds enormous potential for improving people’s health and well-being—and researchers are just getting started.
“I love it,” Hall said. “It’s a great field, and I feel like we’re working toward something that is going to make a difference.”
—This story was originally published by the University of Maryland Department of Cell Biology & Molecular Genetics
REU Bioinformatics Program Immerses Students into Interdisciplinary Research

A dozen students from around the country are gaining invaluable research skills in bioinformatics this summer at the University of Maryland (UMD) as part of a Research Experience for Undergraduates (REU) program.
Sponsored by the National Science Foundation, the Bioinformatics Research in Data Science for Genomics (BRIDGE) REU exposes visiting undergraduates to interdisciplinary training needed to process and analyze large scale genomic datasets.
The hands-on program places students in pairs, partnering them with a faculty mentor for a 10-week research project that concludes on August 9. Each student receives a weekly stipend, room and board, and the opportunity to collaborate with some of the university’s top experts in computational biology.
“We want to give them an opportunity to learn what research is about, so they can decide if they want to follow a research career,” says Mihai Pop, a professor of computer science and director of the University of Maryland Institute for Advanced Computer Studies (UMIACS). “We're giving them the experience of what it’s like to do authentic research.”
Pop is one of several faculty in the Center for Bioinformatics and Computational Biology (CBCB) that are active in this year’s REU.
He is working with students and others to identify phage plasmids. Phages are a type of virus that infect the bacterium, inserting themselves in the bacterium genome and potentially reproducing and killing the cell.
Plasmids are another structure in chromosomes that exist within bacterial cells that are known for carrying antibiotic resistance. Some phages masquerade as plasmids when they infect bacteria.
There is very little information available on the process of a phage morphing into a plasmid, so Pop’s group is doing an analysis on bacterial communities from the human gut to learn more.
“Students are trying to classify pieces of DNA that we think look like plasmids and decide whether those are just plasmids, or they are phage plasmids,” explains Pop, who is leading this year’s BRIDGE cohort at UMD.
Seth Commichaux, who is currently a bioinformatics research scientist at the U.S. Food and Drug Administration (FDA), is also active in this year’s BRIDGE program, mentoring a pair of students that are also looking into plasmids.
“Where I currently work, I don't have much access to students. Most of my interactions are with people either at my level or more senior,” says Commichaux, who graduated from UMD in 2022 with a doctoral degree in computational biology. “Being able to mentor skilled undergraduates is something that I really enjoy, and it's been very rewarding.”
Sahana Rangarajan is one of two students working with Commichaux. Rangarajan, who is visiting from the University of California San Diego, had done prior work involving bacteriophages. Determining the host range of plasmids and bacteriophages involve similar logic, according to Rangarajan.
James Mwihaki, a computer science major at MidAmerica Nazarene in Kansas, is the other student being mentored by Commichaux.
The ability to bring their past research experience and apply it to this REU is something both Rangarajan and Mwihaki say they value.
“I’m able to draw from my background a lot to inform the direction we’re going with in terms of how we’re approaching and what biological features we’re going to be working with,” says Rangarajan.
Michael Cummings, a professor of biology and the director of the CBCB, is part of a group of researchers partnering with a local drug rehabilitation center to identify trends that can translate into tailored treatment plans and improve recovery rates for people struggling with addiction.
Cummings (pictured left) is working this summer with several REU students on that project, applying machine learning algorithms to electronic medical record data to measure various patient experiences and outcomes.
Ryan Schuenke (pictured right), who is entering his senior year at Drake University, is collaborating with Cummings on the project. Schuenke says the program hasn’t been exactly what he envisioned, but has proven invaluable, nonetheless. 
“It filled in a lot of the questions I had regarding high-level research, and it reassured me that research is what I want to do,” he says.
There are two other BRIDGE research projects this summer led by CBCB faculty.
Heng Huang, a Brendan Iribe Endowed Professor in Computer Science with an appointment in UMIACS, aims to test the relationship between single-cell RNA sequences and the genetic information of the cell. His REU team hopes to build a machine learning model to predict the phylogenetic information from RNA count data.
Rob Patro, an associate professor of computer science with an appointment in UMIACS, is leading an REU group developing a sequence search engine over the CHESS database, which contains 27 million identified human genomic transcripts. The REU team is hoping their system will avoid re-discovery of transcripts that are unlikely to be functional, as well as provide potential evidence that previously identified database sequences may have a meaningful biological function.
— Story by Shaun Chornobroff, UMIACS communications group
In 2024, there are three other REU’s active on the Maryland campus in addition to the one focused on bioinformatics. Bill Gasarch, a professor of computer science, leads an REU focused on machine learning and AI, among other topics. Maria Cameron, a professor of mathematics and member of the UMD Center for Machine Learning, is leading an REU on modern topics in pure and applied mathematics; and the Institute for Trustworthy AI in Law & Society is running an REU focused on participatory design, methods and metrics, and evaluating trust.
Patro Awarded $400K to Enhance Open-Source Gene Expression Tools
The concept of gene expression analysis might compare to an orchestra’s conductor—a guiding force for researchers intent on deciphering the genomic melodies within our cells. From unraveling the mysteries of diseases to better understanding human development, this scientific method significantly advances biomedical research by shedding light on life’s inner workings.
To further research and scholarship that is focused on gene expression analysis, the Chan Zuckerberg Initiative (CZI) has awarded a $400,000 grant to a team of University of Maryland researchers. CZI is a philanthropic effort launched in 2015 by Meta founder Mark Zuckerberg and his wife, Priscilla Chan.
The award supports efforts by Rob Patro, an associate professor of computer science, in unifying and enhancing open-source transcriptomics tools—freely available software programs designed to analyze RNA transcripts, enabling researchers to efficiently study gene expression and regulation.
Patro, who has an appointment in the University of Maryland Institute for Advanced Computer Studies (UMIACS), says he is honored to receive this grant, saying it highlights the value of his lab’s commitment to furthering open science methodology and expanding access to open-source, practical and robust scientific software.
“This award will improve upon the core algorithms and data structures of software tools that have been used in thousands of gene expression analysis studies to date, increasing performance and reducing runtime and resource requirements and hence, costs,” says Patro, who is a core member of the Center for Bioinformatics and Computational Biology. “At the same time, it will build new bridges of interoperability between popular tools, accelerating existing pipelines and providing scientists with more choices in how best to analyze their data.”
One primary goal, Patro says, is to enhance the use and impact of a popular open-source tool known as the STAR aligner, which uses a suffix array—a data structure for efficiently handling large sequences of DNA, RNA or protein data—to align genomic sequencing data.
Despite the suffix array’s long history and optimization for efficient construction, it hasn’t fully utilized modern hardware capabilities, Patro says.
Working closely with Jamshed Khan, who just completed his fifth-year as a computer science doctoral student—and in collaboration with UMIACS colleagues Laxman Dhulipala and Erin Molloy as well as graduate student Tobias Rubel—the UMD team has developed a new parallel algorithm for suffix array construction that outperforms existing methods, building the index faster while simultaneously reducing memory usage. By bringing this novel algorithm and other enhancements to the STAR aligner, the team hopes to improve gene analysis tool’s performance.
Another focus is to integrate various transcriptome analysis tools. The UMD researchers will create a high-bandwidth connection between STAR and their own Salmon tool for transcript quantification from bulk RNA sequencing data, reducing runtime and resource usage. A similar integration will be built between STAR and their alevin-fry tool for single-cell gene expression analysis.
Lastly, along with Daniel Liu (a collaborator and recent graduate from UCLA) and Noah Cape (a student at Williams College whom Patro advised as part of the CBCB’s REU program), Patro is developing a universal adaptor tool for single-cell sequencing analysis, accommodating different data formats and protocols. This tool will enable existing single-cell expression quantification tools to process diverse types of data without requiring code modifications for each new input format.
Patro’s winning proposal was one of 32 that were funded this year through CZI’s Essential Open-Source Software for Science program, which supports open-source software projects that are essential to biomedical research. Its goal is to support software maintenance, growth, development and community engagement for these critical tools. This year’s program co-funders are the Kavli Foundation and the Wellcome Trust.
This marks the second time that Patro has received funding from CZI. In 2022, he was awarded $350K to improve upon a collection of interrelated tools his lab developed to process genomic data.
—Story by Melissa Brachfeld, UMIACS communications group
Cummings Employs Machine Learning to Tackle Addiction Crisis

Baltimore is among the epicenters of the nationwide substance abuse crisis. The city was recently named the “Overdose capital of the U.S” after a joint investigation by The New York Times and Baltimore Banner found it averaged 1,000 overdose deaths per year, a death rate nearly double of any other large American city.
In a concerted effort to curtail overdose deaths and improve treatment for people struggling with substance abuse, researchers at the University of Maryland (UMD) and University of Maryland, Baltimore (UMB) have partnered with a regional drug treatment center to apply machine-learning techniques to electronic medical record (EMR) data.
By using EMR to analyze data from patients including race, gender, location, traumas suffered, educational background and income, the cross-institutional team is hoping to identify trends that allow treatment to be better tailored to the individual and improve recovery rates.
“Our complex histories, individual abilities and family situations are all very personal to us, but there's going to be some commonalities among people suffering from addiction,” says Michael Cummings, a professor of biology with an appointment in the University of Maryland Institute for Advanced Computer Studies (UMIACS) who is one of the researchers on the project.
Cummings’ partners on the project include Edward Bernat, an associate professor of psychology at UMD, and Fadia Shaya, a Distinguished University Professor in the school of pharmacy at UMB. Yi Chen, a fifth-year doctoral student in UMD’s biological sciences program advised by Cummings, is also involved with the project.
Their work is supported by a $50,000 grant from the Institute for Clinical and Translational Research and the University of Maryland, Baltimore.
Bernat has extensively researched substance use disorders and its intersection with psychopathology. For the past few years, he has been working directly with Tuerk House— an inpatient drug center and crisis stabilization center in West Baltimore that sees 500–600 patients a month. Bernat provides ongoing clinical intake diagnostic services for the center’s clinical team.
Not long after he began conducting evaluations of the EMRs for patients arriving at Tuerk House, Bernat saw a need to involve machine learning to get the most out of the data he was viewing. This was due to the larger amount of data, from a larger number of patients.
“That’s where machine learning often has a big advantage—going through all of those different variables, and getting it crunched down into what’s actually meaningful with the minimal data available,” says Bernat.
The researchers plan to run machine learning algorithms on up to 10,000 of Tuerk House’s unique intake records from 2022 and 2023, with a focus on additional hurdles that Black substance users face that can impact their path to recovery.
African Americans complete substance abuse treatment only 40% of the time, which is 10% less than their White counterparts, according to a 2016 study published in the Journal of Substance Abuse Treatment. The study said that factors like socioeconomic status— specifically high school attainment and employment—are both associated with higher completion rates.
“If we find some subpopulations of people with certain characteristics are more at risk for relapse or something like that, then we can modify treatment for those sets of individuals.,” Cummings says.
Tuerk House’s partnership with UMD researchers for this project is part of a broader developing collaborative relationship to advance health disparities and treatment research relevant to Black substance users.
The inpatient drug center is in a majority black city that has been disproportionately damaged by the addiction epidemic, one that Chen is incredibly familiar with.
Chen moved to Baltimore to attend Johns Hopkins University, where he completed his B.A. in biology in 2014. For four years he did community service at Stepping Stone Ministry, a faith-based organization on the Hopkins campus that gave aid to the local homeless community, only stopping in 2020, when COVID–19 forced him to do so.
“Substance use disorder and homelessness are highly comorbid,” Chen says. “So, in some ways, this project is another way of serving the same population from that part of my life using a different set of skills.”
This project contrasts Chen’s previous work which has centered around molecular biological data sets, posing occasional challenges in refining and understanding data.
Chen noted the significance of the collaboration of experts from across various fields for this project.
“Substance use disorder is a complex disorder,” he says. “This project can hopefully address that by virtue of being a collaborative work involving psychopathology, social determinants, and machine learning.”
Looking ahead, Bernat envisions a dashboard wherein a patient completes the intake assessment at Tuerk House, and the system the research team is building will suggest individualized treatment plans based on the data received.
“Our local goal with is to develop something with [Tuerk House] clinicians that they can use quickly and efficiently,” he says.
Bernat and Cummings both emphasized that the overall goal is to understand different populations and improve the success rate of addiction treatment through a data-driven, personalized approach.
“That’s the ultimate benchmark we’re looking to achieve,” says Cummings. “Hopefully, data science and machine learning can provide some insights on how specialized health care can be improved and become more individualistic.”
— Story By Shaun Chornobroff, UMIACS communications group
Using Machine Learning to Develop a More Effective Malaria Vaccine
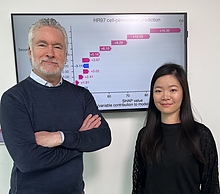
Caption: Michael Cummings (left) and Renee Ti Chou (right) collaborated on the project with researchers from the University of Maryland School of Medicine.
While cases of malaria have been decreasing in some parts of the world thanks to drug-based therapies, efforts to eradicate this life-threatening disease have recently stalled, due in part to the parasites that transmit malaria becoming resistant to current therapeutics.
Computational biologists from the University of Maryland have partnered with researchers at the University of Maryland School of Medicine to address this challenge. Using a novel approach called reverse vaccinology, which employs powerful bioinformatic tools and reverse pharmacology practices, the researchers are examining the genetic makeup of several parasites that cause malaria, seeking specific antigens to target with new vaccines.
The team’s initial research was just published in npj Systems Biology and Applications, a leading journal that focuses on scientific work that takes a systems-based approach.
The paper’s lead author is Renee Ti Chou, a data scientist at Lexical Intelligence in Rockville, Maryland, who earned her Ph.D. in computational biology last year at the University of Maryland.
Co-authors include Michael Cummings, a professor of biology with an appointment in the University of Maryland Institute for Advanced Computer Studies, and Amed Ouattara, Matthew Adams, Andrea A. Berry and Shannon Takala Harrison, all from the University of Maryland School of Medicine’s Center for Vaccine Development and Global Health.
Without a good vaccine, the researchers say, it is hard to get rid of any disease, malaria included. But to make a truly effective vaccine for malaria—which killed an estimated 608,000 people in 85 countries in 2022—scientists need to target different life cycle stages of the P. falciparum parasite, the deadliest species of Plasmodium that causes malaria in humans.
This can be tricky, though, because not only does the parasite take on different forms during its life cycle, but its genetic proteins also change, making it difficult for the human immune system to counter. So far, most vaccine efforts have focused on a few proteins without looking at the whole picture of the P. falciparum parasite’s genes.
With the reverse vaccinology approach, based in-part on powerful machine learning algorithms, the UMD/UMB researchers hope to gain a clearer picture that will lead to better vaccine antigens.
For their work, the researchers analyzed thousands of proteins from the P. falciparum parasite, considering 272 different factors for each. They used a machine learning technique called positive-unlabeled learning to sort through this data, focusing on proteins that resemble known vaccine targets.
They do not have to start with specific criteria; instead, they let the computer learn from what is already known about effective vaccine targets. This method looks at all the genes of the malaria parasite to find ones that might make good vaccine targets, even if they are not obvious at first.
Cummings, who is also the director of the Center for Bioinformatics and Computational Biology, says this approach has not been widely used for malaria, but it could help find new vaccine candidates, especially for parts of the P. falciparum parasite that are not well understood yet.
The researchers not only identified new potential vaccine targets, Cummings adds, but also ranked them based on importance. To prioritize the most promising candidates, they looked at factors like gene essentiality and when the proteins are active in the parasite’s life cycle.
“These findings offer a flexible framework for future vaccine research,” says Cummings, who was Chou’s academic adviser at UMD. “We can adjust our criteria and even apply this approach to other diseases beyond malaria. It’s a big step forward in the quest for better vaccines.”
The team’s research is supported by the National Science Foundation, the National Institutes of Health (NIH), a National Health and Medical Research Council grant, and a grant from MPowering Maryland. Other support came in the form of the Anne G. Wylie Dissertation Fellowship, which Chou received in her final year of Ph.D. training at UMD.
For her doctoral work related to this project, Chou recently received the Charles A. Caramello Distinguished Dissertation Award. Also, in work that is peripherally tied to this current research, Cummings was the recipient of an NIH grant in 2023 to study the body’s immune response to malaria to help scientists develop more effective vaccines.
—Story by Melissa Brachfeld, UMIACS communications group
The New Frontier of Microbiome Science: Computational Challenges and Solutions
The microbiome refers to the whole sum of microorganisms in a particular environment, such as the collective sum of gut bacteria in a human being. Microbiome research is a new frontier of scientific exploration. Studies that use big data technology to examine whole genomes of hundreds of organisms simultaneously represent a field called metagenomics. As this field matures, scientists are increasingly recognizing the need for sophisticated tools and technologies to decipher the complexities hidden within these microbial ecosystems.
To that end, on April 2, Mihai Pop, a professor in the Department of Computer Science and the director of the Institute for Advanced Computer Studies at the University of Maryland, gave a talk on the analytical challenges of microbiome science and how they can be combated by computational methods. The talk focused on the pivotal role of computational tools in unraveling the secrets of microbiomes and addressing the challenges associated with analyzing the vast datasets generated by these studies.
A key focus of metagenomics is the taxonomic classification of different microbes. The primary method for organizing and classifying microbes is comparing them to a database of known organism sequences. These similarity-based techniques are especially effective when the organisms in the sample are well represented in the database. Pop mentioned one of the most common similarity search methods used to classify microorganisms, the Basic Local Alignment Search Tool (BLAST). However, BLAST often misidentifies the closest neighbor to the microorganism of interest; the “most similar” organism according to BLAST may not actually be the most closely related.
“How can we find what’s the real [closest hit] if there is a hit? The E-value is misleading,” Pop explained during the talk, suggesting that BLAST may not always accurately identify the most similar organism to the microbiome of interest.
The E-values Pop mentioned refer to parameters in BLAST that describe the number of hits one can “expect” to see by chance when searching a database of a particular size. Pop also emphasized how many of these problems were only discovered years after BLAST integrated into common use.
“These are things we found out 20, 30, 40 years after [the computational tool] was written ... even though something has been used for many, many years, there [are] still things to learn about it,” Pop explained.
One of the other main challenges Pop highlighted is how the structure of biological databases affects scientists’ ability to reliably reveal insights on the microbiome. Reference databases are not all-encompassing. Many microorganisms cannot be cultured in labs, and a large proportion of those that can have not been sequenced or added to these reference databases. Consequently, not all environmental organisms are included in the sequence database, which limits the accuracy of similarity-based methods.
These problems are further compounded by the lack of contiguous information available in most sequencing datasets. Many sequencing analyses have to begin by joining together many sequence fragments and stitching together a whole related sequence. Assembling the sequencing data is also an unstandardized process, as new technologies used for assembling genomes are constantly being developed. These limitations can impede researchers' ability to derive meaningful insights and connections from microbiome datasets, because it substantially limits precision and decreases the accuracy of reference databases.
Pop then transitioned to discussing algorithms and software approaches to sequence similarity. Many current software used in classification employ the most recent common ancestor (MRCA) method. MRCA provides an annotation (marking of a specific feature of the DNA sequence) at the broadest taxonomic class that encompasses all of the possible markings in a sequence. However, this means that different types of software that use MRCA only make a few classifications at the genus or species level, meaning that stronger relationships between two microbes cannot be determined at the family, class or phylum level.
To address this challenge, Pop shared efforts from his own lab to develop advanced computational tools tailored specifically for microbiome analyses. He specifically focused on the Ambiguous Taxonomy eLucidation by Apportionment of Sequences (ATLAS). ATLAS is a data-driven database partitioning method, which aims to divide a large dataset into smaller, more easily analyzable datasets. ATLAS groups sequences into biologically meaningful partitions by querying the sequence against a reference database and then identifying and clustering hits that are considered significant. ATLAS also represents a shift away from the MRCA method.
As the talk concluded, Pop emphasized the critical need for interdisciplinary collaboration to advance microbiome research. Integrating expertise from fields such as biology, computer science and statistics is essential for developing innovative solutions to microbiome-related challenges. This interdisciplinary approach allows researchers to harness the power of computational tools to extract meaningful patterns and associations from microbiome datasets.
—This article by Shreya Tiwari was originally published in The News-Letter, a student publication at John Hopkins University.
Marking Five Years of Research, Scholarship and Innovation in the Iribe Center

A well-designed building is more than just a structure—it’s also a catalyst for collaboration, fostering a dynamic environment where people come together to brainstorm ideas and create new technologies.
The Brendan Iribe Center for Computer Science and Engineering is exactly such a place. Launched five years ago this month, the 215,000 square foot structure is home to the Department of Computer Science and the University of Maryland Institute for Advanced Computer Studies (UMIACS).
A visit to the ground floor of Iribe offers multiple experiences. Computer science is the largest major on campus—and one of the largest in the nation—with visitors keenly aware of this fact as they observe hundreds of undergraduates heading to class or studying in the expansive lobby area.
Cutting-edge research is also visible on the ground level, with a shiny silver mannequin staring at you from the Small Artifacts Lab (it’s used to test wearable technologies), miniature drones buzzing in the Brin Family Aerial Robotics Lab, and digital art or other 3D visuals captivating your attention as you walk by the Immersive Media Design lab space.
For UMIACS, the Iribe Center is a boon for the seamless flow of bold ideas and new knowledge across multiple academic disciplines.
The institute currently supports more than 80 faculty and 200 Ph.D. students from 15 departments on the UMD campus. Most of these faculty and students reside in the Iribe Center, spread out in offices, labs and workspaces on the third and fourth floor of the building as well as part of the fifth floor that’s home to the Maryland Cybersecurity Center.
“Most scientific advances that have transformed our world have emerged at the boundary between disciplines,” says Mihai Pop, a professor of computer science who is the director of UMIACS. “Our continued scientific leadership critically depends on scientists from multiple disciplines coming together to work on impactful projects.”
For Naomi Feldman, a professor of linguistics, the Iribe Center has offered abundant opportunities to interact with faculty she might not normally see in her home department.
“I’m a computational linguist and having colleagues literally right down the hallway working on computational acoustics, computer vision and machine learning has been very advantageous for me and for my students,” Feldman says.
The building has tremendous aesthetic value as well, faculty members say.
Vanessa Frias-Martinez, an associate professor in the College of Information Studies, says the airiness of the Iribe Center and the stunning views it offers make for a calm and creative workspace. 
“My office has large windows and plenty of natural light, and we have our beautiful rooftop garden—both serve as a source of inspiration and comfort,” says Frias-Martinez, who is director of the Computational Linguistics and Information Processing (CLIP) Lab. “I feel like I can really focus on my work and be productive, but it’s also nice to pause and unwind with the fantastic views and green spaces we have available.”
Feldman, also a member of the CLIP Lab, says that the Iribe Center has multiple spaces for students to meet and network and for faculty to host workshops and seminars.
“Our students come from diverse academic backgrounds—linguistics, computer science, information studies, human-computer interaction, and more—and the workshops and talks in the Iribe Center offer a robust opportunity to communicate across disciplines,” Feldman says.
Michael Cummings, a professor of biology who leads the Center for Bioinformatics and Computational Biology, says that in addition to his graduate students having a modern, inviting workspace, moving to the Iribe Center has made many of his day-to-day administrative tasks easier and more productive.
“I’m able to walk down the hall and talk with the business office on the status of a research grant or meet with our tech staff to go over our computing needs,” Cummings says. “This is not only more efficient, but also more pleasant.”
With research and innovation in UMIACS constantly expanding, the Iribe Center will continue to serve its mission, and serve it well, says UMIACS director Pop.
“We’re thankful for the private donations and state support that made this building possible,” Pop says. “But it remains up to the people that work here—faculty, students and staff—to fulfill the vision of why this space was built: we’re a hub for technology, collaboration and discovery that will have an impact on the world.”
—Reporting on this story by Melissa Brachfeld, UMIACS communications group
